U-Net
Architecture
The architecture in UNET consists of two paths. Contracting path (encoder network) followed by an expansive path (decoder network). U-Net gets its name from the architecture, like the letter U, as depicted in the image below.
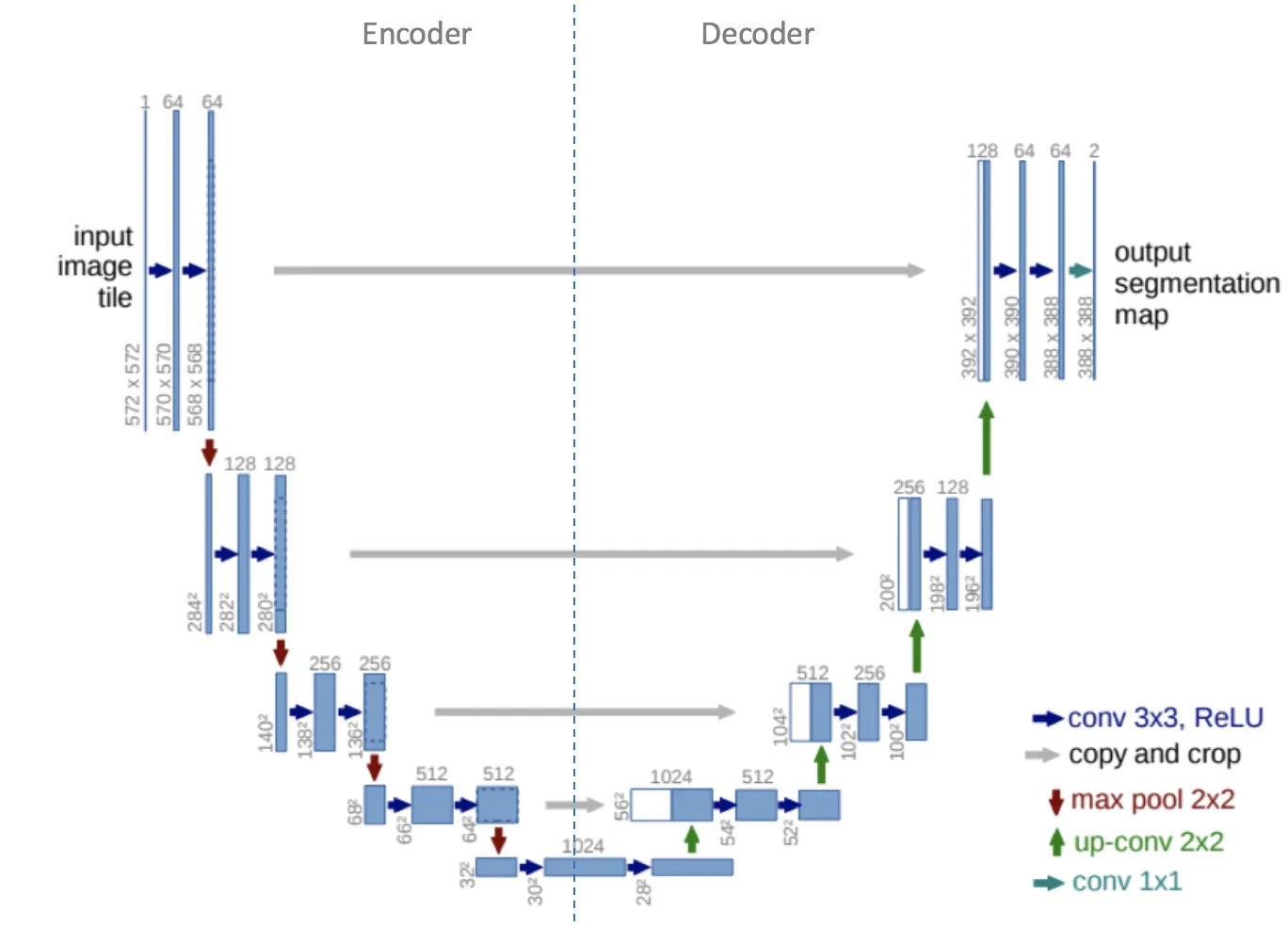
The encoder (left side) typically consists of a pre-trained classification network, such as ResNet, using convolution blocks followed by maxpool downsampling to encode the input image into feature representations at various levels. In this architecture, there is repeated application of two 3x3 convolutions. Each convolution is followed by a ReLU and batch normalization. Then a 2x2 max pooling operation is applied to reduce dimensions by half. Again, at each downsampling step, we double the number of feature channels, while we cut in half the spatial dimensions.
The decoder path (right side) consists of upsampling of the feature map followed by a 2x2 transpose convolution, which cuts the number of feature channels in half. The concatenation with the matching feature map from the contracting path, as well as a 3x3 convolutional filter (each followed by a ReLU) is also implemented. A 1x1 convolution is employed in the final layer to map the channels to the required number of classes.
Code Implementation example
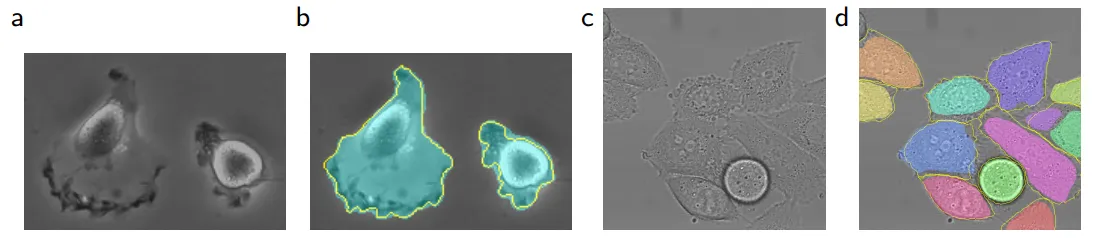
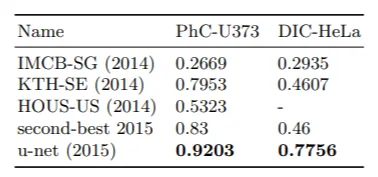
U-Net Segmentation results (IOU) on PhC-U373 and DIC-HeLa datasets.
Parameters in model playground
Encoder network
In Model Playground, we can select feature extraction (encoding) network to use as either ResNet or EffiecientNet. The depth of the ResNet model/ EfficientNet sub type has to be specified as well.
Weights
After selecting the desired encoder network, the weights to be used for model initialization can be chosen.
Further Resources
U-Net: Convolutional Networks for Biomedical Image Segmentation
